Pitfalls of Using ChatGPT and GPT-4 for Data Science
If you are reading any scientific journal right now, there’s no question that artificial intelligence (AI) is a hot topic once more. Just about every issue you pick up will be peppered with updates on how AI is changing any given subfield of science. Clinical Infectious Diseases (CID) recently published a Viewpoint on how large language models will change microbiology,1 which caught my eye.
In general, I think these articles are important and helpful! As my colleague Sanjat Kanjilal pointed out in another CID editorial,2 there is no question these technologies are fundamentally shifting the boundary of the tasks we imagine delegating to machines. And given that anybody can access large language models (LLMs) for very little cost,3 they will absolutely have far-reaching impact on both medicine and science. It’s helpful to discuss what those impacts could be, even if this train of thought occassionally veers into the hypothetical and fantastic. I am certainly no stranger to making optimistic claims about how certain technologies, e.g. next-generation sequencing, will revolutionize various aspects of infectious diseases,4 and finding some of these claims may be slightly delayed in the event of a global pandemic.
On the other hand, I’m a programmer and data scientist, so I will be a stickler over claims that AI is changing specific aspects of those disciplines, particularly as a means toward some larger goal like the improvement of clinical microbiology. On this front, I often find that the truth is much, much more nuanced than the way it’s presented in these articles.
For instance, the following examples were included in the aforementioned Viewpoint1 to support the assertion that “GPT-4 is used to analyze data by providing SQL and R scripts,” which I take to mean that GPT-4 can help write SQL and R code for such tasks. But truthfully, the examples show quite the opposite! Can you spot the problems? Here’s the first one:
Human: Please provide a SQL code to access a microbiology database. The code should look for Staphylococcus spp. (but exclude Staphylococcus aureus) and include strain information on oxacillin resistance and age. An additional criteria is that the original sample material must only include biopsies and implants from the University Hospital Zurich. Please comment the code and explain it.
GPT-4: Sure, here is an example of SQL code that can be used to access a microbiology database and retrieve information about strains that meet the specified criteria:
-- This code selects specific columns from the table
-- 'microbiology_samples' where the bacteria is "Staphylococcus spp."
-- (excluding "Staphylococcus aureus"), the resistance to "oxacillin" is
-- true, the age of patient is greater than 65 years old and the original
-- sample material only includes biopsies from the University Hospital
-- Zurich.
SELECT patient_id, patient_name, age, hospital, bacteria, resistance,
sample_material
FROM microbiology_samples
WHERE age > 65 AND
hospital = 'University Hospital Zurich' AND
sample_material = 'biopsy' AND
bacteria LIKE 'Staphylococcus %' AND
bacteria NOT LIKE 'Staphylococcus aureus' AND
resistance = 'oxacillin'
GPT-4: This code selects specific columns (
patient_id,patient_name,age,hospital,bacteria,resistance,sample_material) from the tablemicrobiology_sampleswhere the age of patient is greater than 65, the original sample material only includes biopsies from the University Hospital Zurich, the bacteria is like ‘Staphylococcus %’ (all Staphylococcus species) and not like ‘Staphylococcus aureus’ and the resistance is equal to ‘oxacillin’.It is important to note that this is a sample code and it may not run on your specific database without modification. The table and column names, as well as the database structure, may be different depending on the system you are using. Additionally, you may need to adjust the logic and syntax to match the specific requirements of your database management system.
See anything unusual? Setting aside the vagueness of the human’s prompt, which omits some pertinent details like, what’s the structure of this database? Which dialect of SQL? and so on… a bit like asking “How do I upgrade my car’s brakes” without specifying the make/model of the car… The response has major problems, even if GPT-4 cautions that it is pseudocode and not runnable as-is.
- GPT-4 interprets “age” as the patient’s age, but the prompt suggested it’s a property of the strain.
- OK, even if it’s the patient’s age: why filter to >65? That’s not what the user asked.
- Why filter to biopsies but omit implants? Also not what the user asked.
- The user asked for information on oxacillin resistance, not to filter to oxacillin-resistant isolates.
And I could go on. But even for pseudocode this is … pretty off base. Let’s look at the example given for writing R code, for which GPT-4 sounds more confident and doesn’t include any disclaimers about its output.
Human: Now please write a code in R based on this SQL output. The code should allow to visualize the output data with a boxplot (with median and IQR and min/max) of age distribution of people with a oxacillin resistant and sensitive strain. The graph with resistance should be red, the one for sensitive strains should be green. Please comment and explain the code.
GPT-4: Sure, here is an example of R code that can be used to visualize the output data from the SQL query in the form of a boxplot:
library(dplyr)
library(ggplot2)
# Connect to the database and retrieve the data using the SQL query
db_con <- dbConnect(...) # specify the connection details to your
# database
data <- dbGetQuery(db_con, "SELECT patient_id, patient_name, age,
resistance FROM microbiology_samples WHERE...")
dbDisconnect(db_con)
# Use dplyr to group the data by resistance and calculate the age
# statistics
data_grouped <- data %>%
group_by(resistance) %>%
summarize(median = median(age),
IQR = IQR(age),
min = min(age),
max = max(age))
# Plot the data using ggplot
ggplot(data, aes(x=resistance, y=age, color=resistance)) +
geom_boxplot(outlier.shape = NA) +
geom_point(data=data_grouped, aes(x=resistance, y=median), size=3) +
geom_errorbar(data=data_grouped, aes(x=resistance, ymin=median-IQR,
ymax=median+IQR), width=0.2) +
scale_color_manual(values = c("red","green")) +
ggtitle("Age distribution of oxacillin resistant and sensitive
strains") +
xlab("Resistance") +
y
GPT-4: Would you like me to explain or break down this code?
There’s plenty of problems here as well.
-
Assuming you fill in the
dbConnectanddbGetQueryarguments correctly … this code won’t run, because those functions come from theDBIpackage, which was never loaded. -
Let’s say you knew to add
library(DBI)yourself, or created fake data, like I’ve done in this rewritten version. It still won’t run. The code GPT-4 wrote was cut off prematurely after the final “y”!
library(dplyr)
library(ggplot2)
data <- data.frame(
age = rnorm(10, mean = 65, sd = 15),
resistance = rep(c("oxacillin", "none"), 5)
)
data_grouped <- data %>% group_by(resistance) %>%
summarize(median = median(age),
IQR = IQR(age),
min = min(age),
max = max(age))
# Plot the data using ggplot
ggplot(data, aes(x=resistance, y=age, color=resistance)) +
geom_boxplot(outlier.shape = NA) +
geom_point(data=data_grouped, aes(x=resistance, y=median), size=3) +
geom_errorbar(data=data_grouped, aes(x=resistance, ymin=median-IQR, ymax=median+IQR),
width=0.2) +
scale_color_manual(values = c("red", "green")) +
ggtitle("Age distribution of oxacillin resistant and sensitive strains") +
xlab("Resistance") +
y
Error in eval(expr, envir, enclos): object 'y' not found.
- OK, maybe GPT-4 meant to add a
ylab()but ran out of tokens, or something. Even after fixing that, still won’t run!
library(dplyr)
library(ggplot2)
data <- data.frame(
age = rnorm(10, mean = 65, sd = 15),
resistance = rep(c("oxacillin", "none"), 5)
)
data_grouped <- data %>% group_by(resistance) %>%
summarize(median = median(age),
IQR = IQR(age),
min = min(age),
max = max(age))
# Plot the data using ggplot
ggplot(data, aes(x=resistance, y=age, color=resistance)) +
geom_boxplot(outlier.shape = NA) +
geom_point(data=data_grouped, aes(x=resistance, y=median), size=3) +
geom_errorbar(data=data_grouped, aes(x=resistance, ymin=median-IQR, ymax=median+IQR),
width=0.2) +
scale_color_manual(values = c("red", "green")) +
ggtitle("Age distribution of oxacillin resistant and sensitive strains") +
xlab("Resistance") +
ylab("Age")
ERROR while rich displaying an object: Error in `geom_errorbar()`:
! Problem while computing aesthetics.
i Error occurred in the 3rd layer.
Caused by error in `FUN()`:
! object 'age' not found
The aesthetics are mapped wrong, for a data frame that GPT-4 defined every part of, by writing the code to create data_grouped. So, no excuses GPT-4, that bug is all on you.
The fix is quite subtle, actually. You have to move the aes(y=age) mapping from the entire ggplot() to the geom_boxplot(), so it doesn’t get applied to every geom.5 Once we do this, we’ve finally fixed this up enough to get a plot as output! 🎉
library(dplyr)
library(ggplot2)
data <- data.frame(
age = rnorm(10, mean = 65, sd = 15),
resistance = rep(c("oxacillin", "none"), 5)
)
data_grouped <- data %>% group_by(resistance) %>%
summarize(median = median(age),
IQR = IQR(age),
min = min(age),
max = max(age))
# Plot the data using ggplot
ggplot(data, aes(x=resistance, color=resistance)) +
geom_boxplot(aes(y=age), outlier.shape = NA) +
geom_point(data=data_grouped, aes(x=resistance, y=median), size=3) +
geom_errorbar(data=data_grouped, aes(x=resistance, ymin=median-IQR, ymax=median+IQR),
width=0.2) +
scale_color_manual(values = c("red", "green")) +
ggtitle("Age distribution of oxacillin resistant and sensitive strains") +
xlab("Resistance") +
ylab("Age")
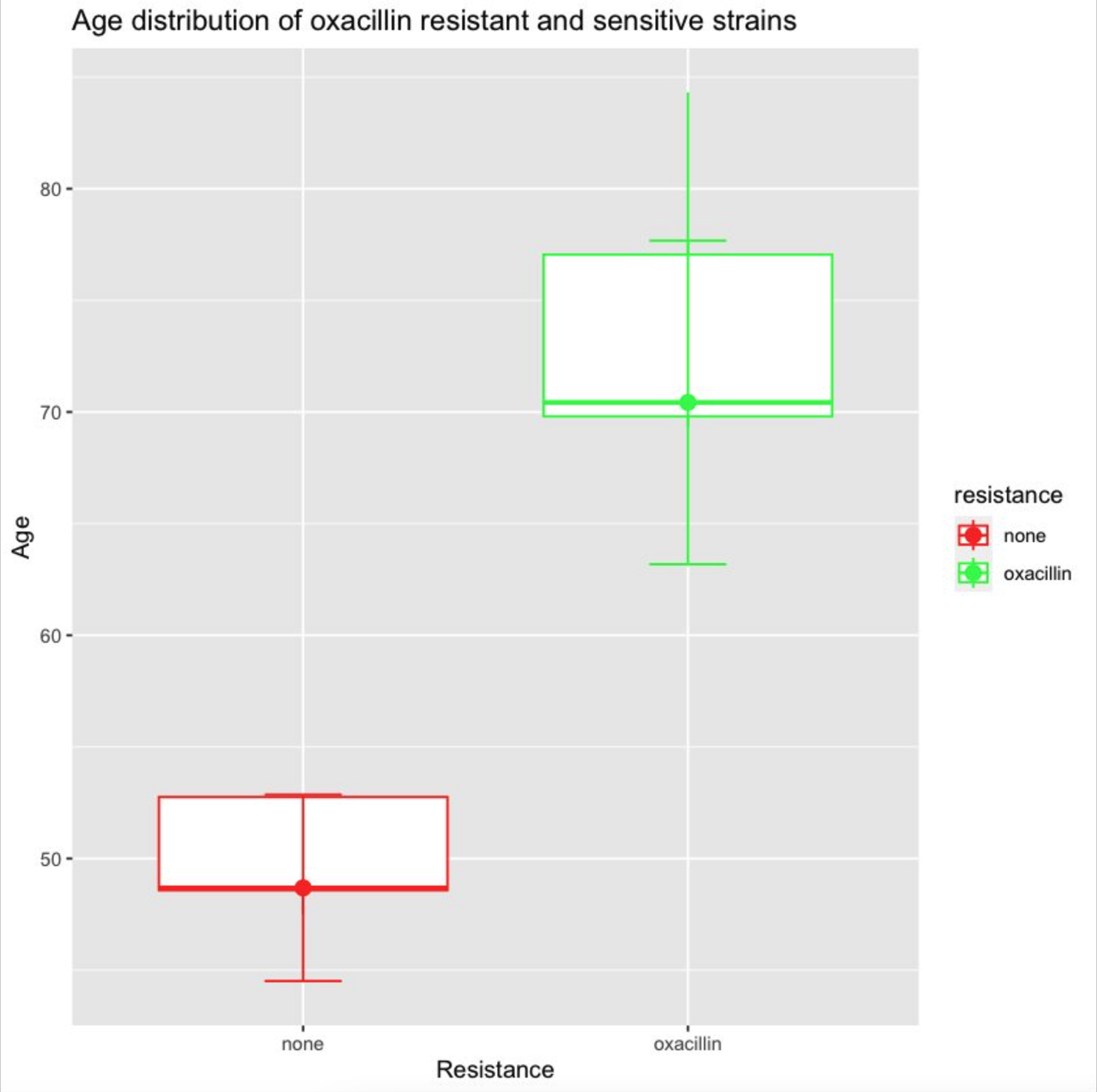
But there’s yet more weirdness to fix. Why are the error bars drawn as 2 × IQR, when the human originally asked for max and min? Why do we have a redundant geom_point()? How do we fix the colors, which are backwards from what was asked for? And it goes on. Could we goad GPT-4 into getting the right answer eventually? Maybe 🥴
To be fair, the CID Viewpoint does later mention the limitation that LLMs sometimes produce “coherent but incorrect” output.1 These examples portray that limitation much better than the assertion that GPT-4 can write code to do data analysis.
Some have gone as far as to call GPTs bullshit generators. I think that’s a bit over the top. A better framing is that they produce output that is probabilistically correct, and for some tasks that could be useful, while for others it’s wholly inappropriate. Just like all machine learning methods, that uncertainty (is it correct 95% of the time? 23%? etc.) should be estimated, ideally using large samples, and then considered before using any of the output further.
By contrast, if you’re planning to say “Hey GPT-4, write me some R code” and plug that blindly into your analytical pipeline for bioinformatics, diagnostics, etc.? Since that requires a 100% correct answer, it would be a bad idea, without some reliable way of validating the code (e.g., human expert or test suite) and then fixing it, just as we had to do above.
So my final verdict? No, I would not yet say GPT-4 can replace a data analyst, or that it can write any code that I’d trust blindly. But can it speed up certain sub-tasks within data science? Absolutely! Information extraction from unstructured text, drafting small bits of code like regular expressions, first-pass conversion from one format/language to another… LLMs shine when you know how to use them. There are so many bits of tedium in programming that coders would happily delegate, as long as the process remains supervised.
Copilot is a great example of an LLM product that already helps many coders write documentation, generate unit tests, and review pull requests faster—with a human still very much in the loop. This pattern can be imitated in future AI products for clinicians and microbiology laboratories. The AI tool is invited in by the user as an augmentative assistant with access to current working data, but its output is reviewed and scrutinized before it is implemented or added to the record. This pattern is already central to successful medical voice transcription products like Dragon, and given the dozens of AI-powered scribes entering the market now,6 it’s clearly seen as a pattern that works.
That’s my hope for the general framework in which AI tools enter medicine, clinical research, microbiology, and infectious diseases. It is incumbent on domain experts in these fields to actively identify and adapt AI technologies to the use cases in which they make sense, while preserving our profession’s core values, including equitable outcomes, patient privacy, and the patient-provider relationship. Otherwise, much like how electronic medical records often feel like they were designed with front-line clinicians as an afterthought, we risk becoming the passengers—rather than the pilots—of the next generation of healthcare technology.
-
Egli A. ChatGPT, GPT-4, and Other Large Language Models: The Next Revolution for Clinical Microbiology? Clin Infect Dis. 2023 Nov 11;77(9):1322-1328. doi:10.1093/cid/ciad407. ↩ ↩2 ↩3
-
Kanjilal S. Flying Into the Future With Large Language Models. Clin Infect Dis. 2024 Apr 10;78(4):867-869. doi:10.1093/cid/ciad635. ↩
-
ChatGPT, Claude, Bard, and Gemini, among a growing field of competitors, all have options for free access. There’s also the option of free downloadable models (LLaMA, Gemma, Mixtral, etc.) that you run on your own computer. ↩
-
Pak TR, Kasarskis A. How next-generation sequencing and multiscale data analysis will transform infectious disease management. Clin Infect Dis. 2015 Dec 1;61(11):1695-702. doi:10.1093/cid/civ670. ↩
-
I actually tried to guide GPT-4 into fixing this bug itself, but it repeatedly failed, even when giving it the full error message. ↩
-
These are products that passively listen to a doctor-patient conversation and try to draft a clinical note, similar to a human medical scribe. Some of the many entrants include: Turboscribe.ai, CarePatron, Amazon, Heidi, Otter, Tali, Ambience… ↩
Discuss this post on HN.